
Decoding the Diet of a Local Owl Population
Abstract¶
Populations of owl species are in danger due to declining habitat. Owls play an essential role in maintaining the local rodent population, which aids in preventing the spread of disease. To better understand the local rodent population and its potential for disease transmission, owl pellets were collected from an abandoned building that was soon to undergo renovation in Burke County, NC. Over two years, 12 pellets were dissected, with eight pellets dissected in the first year and the remaining four in the second year. These pellets were dissected under a biological safety cabinet, resulting in 38 samples. Teeth and jawbones were surface sterilized and dried before pulverization. DNA was extracted from dissected rodent incisors utilizing Qiagen DNEasy spin columns. Mammal primers were used to amplify a 400 bp mitochondrial cytochrome b DNA fragment through Polymerase Chain Reaction (PCR). Mitochondrial cytochrome b is the accepted barcode used for genetic identification of mammals. The resulting amplicons were sequenced and aligned to mitochondrial cytochrome b sequences within GenBank’s BLAST database. In the first year of study, ten sequences most closely aligned with Sigmodon hispidus, commonly known as the Hispid Cotton Rat. Other sequencing attempts were either unsuccessful or suggested contamination. In a second round of study, one DNA sequence aligned with Microtus pinetorum, otherwise known as the Woodland Vole. The remaining DNA sequences were too weak to determine a particular species. Sigmodon hispidus is known for transmitting respiratory diseases that are detrimental to human health, while Microtus pinetorum is known to disrupt foundations in human infrastructure. Prior to building renovations, the owls were relocated to a nearby location where they continue to maintain the prey population as determined in this study.
Due to declining habitats and decreasing wilderness ecosystems, owl populations worldwide are struggling for survival. Owls play a critical role in controlling rat and other rodent populations and preventing the spread of disease. A primary factor in declining habitat is construction. In Burke County, the North Carolina School of Science and Mathematics Morganton (NCSSM-Morganton) is currently in a construction zone. One of the historic buildings that is set to be renovated is inhabited by one or more owls. Throughout this study, the prey of these owls are investigated via DNA analysis of their pellets- the undigested parts of their food that are regurgitated through their beak. Processes such as DNA extraction, gel electrophoresis, and polymerase chain reaction (PCR) were used.
Given that owls represent a large bird family with a cosmopolitan distribution, their pellets are often available in several different habitats Wildlife Online, 2024. They present an affordable, effective tool for detecting small mammals and assist in furthering the knowledge of the ecology and phylogeography of the area. Previous studies have confirmed the potential use of pellet bones as a modern DNA source POULAKAKIS et al., 2005. DNA analysis enables the identification of owl prey at the species level, providing more information when compared to morphological analysis alone. It can be hypothesized that the success rate of DNA analysis in this study will be comparable to that found in similar studies: 45-86% Rocha et al., 2015.
Samples in tropical or subtropical regions are easily degraded due to climatic conditions Philips & Dindal, 1979. Additionally, the accelerated DNA degradation of ancient bones has been related to high humidity and temperatures. Morganton, North Carolina, has a humid, subtropical climate, characterized by hot summers and mild winters. This study assesses if bone samples recovered from owl pellets in this area represent a viable source of modern DNA. If the genetic material uncovers species that educate Burke County on its biodiversity and the impact certain prey species have on the environment, additional actions could be taken to preserve these organisms.
Materials and Methods¶
A similar approach outlined in DNA From Owl Pellet Bones Uncovers Hidden Biodiversity was taken in order to identify local rodent species Rocha et al., 2015. Owl pellets were collected on the NCSSM-Morganton campus to access rodent DNA. DNA extraction was accomplished by employing the Qiagen DNeasy Blood and Tissue kit. A fragment of DNA was amplified through PCR to identify the rodent species consumed by the owls. The DNA fragment amplified for the study was mitochondrial cytochrome b, as it is the accepted barcode used for the genetic identification of mammals and there is a vast database of cytochrome b sequences. Amplicons were sent for sequencing, and matches were identified utilizing Genbank’s database. Disposable face masks, gloves, and lab coats were worn when managing DNA samples to avoid contamination.
Sample Collection¶
Owl pellets were collected in an abandoned building on the developing campus of NCSSM-Morganton. Gloves, disposable face masks, and phone flashlights were used to locate and gather the pellets in individual Ziploc bags. The pellets were frozen for one to two weeks.
Dissection and Sterilization¶
Owl pellets were dissected under a biological safety cabinet using forceps. Bones, including jawbones, teeth, and incisors of rodents were isolated. Several bone samples were selected and sterilized to preserve the DNA. Sterilized distilled water and a 10% bleach solution were used to purify the samples. A three-step cleaning procedure was followed to ensure the bone samples were thoroughly decontaminated (suggesting zero presence of contaminants, including human DNA) and available for DNA extraction.
First, sterile deionized water was used to soak bone samples. Next, the bones were soaked with a 10% bleach solution to completely eradicate contamination Rocha et al., 2015. Lastly, sterile deionized water was used to wash the bone samples, eliminating any excess bleach that could potentially affect future procedures on the sample Rocha et al., 2015. A burner and ethanol were utilized to sterilize the forceps between handling each sample, and the samples were then dried. Pieces of aluminum foil were disinfected by irradiation using a goggle sterilization cabinet. The decontaminated surface of the foil was used to wrap and preserve the sterilized bone samples. An additional layer of aluminum foil was wrapped around the samples in preparation for the pulverization process.
Pulverization¶
Double-wrapped samples were placed in mortars and liquid nitrogen was poured over each specimen. Following this, the bone samples were thoroughly crushed to a fine powder. The ground samples were then placed into separate 1.5 mL microtubes.
DNA Extraction¶
The Qiagen DNeasy protocol was followed in order to extract DNA from samples. Pulverized samples were soaked in 180 µL of Buffer ATL to degrade cell membranes and release the DNA present in the cells of the bony powder. Subsequently, 20 µL of Proteinase K was added to each sample to protect the released DNA from destruction by other proteins Astral Scientific, 2024. A vortexer was used to fully integrate the bony powder with the buffer and Proteinase K Figure 1. The microtubes containing the samples were left in a dry-bath incubator overnight at 56°C.
The following day, 200 µL of Buffer AL was added to each sample microtube, vortexed for fifteen seconds, then incubated for ten minutes at 56°C. Afterward, 200 µL of each sample was transferred to a new set of labeled microtubes. This step was followed by adding 100 µL of ethanol to each newly transferred sample. Samples were transferred once again to respective DNeasy spin columns, which were then placed in a centrifuge for one minute at 8000 rpm with the purpose of isolating DNA Figure 1. The resulting waste was discarded and the centrifuge procedure was repeated with the addition of a series of washing buffers (Buffer AW1 and Buffer AW2) in order to cleanse the DNA of other bio-molecular substances Afnan Uda et al., 2020.
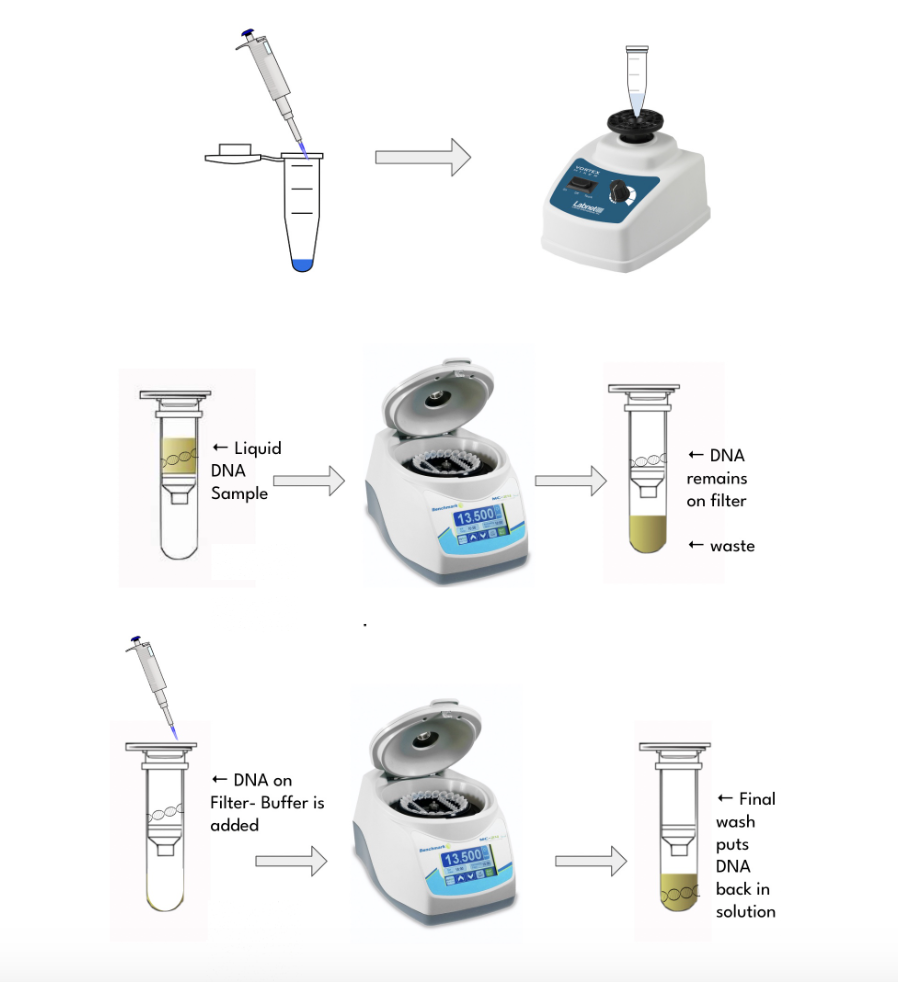
Figure 1:A series of buffers were mixed with samples to disintegrate cell membranes and wash and access DNA. A centrifuge was utilized in order to isolate the clean DNA.
DNA Amplification¶
PCR was employed to execute DNA amplification, the process of duplicating a specific section of DNA, and to help distinguish a particular group of organisms from other groups. For the purpose of the study, mitochondrial cytochrome b, a gene that serves as the genetic insignia for mammals, was amplified. First, 48 µL of a master mix comprising Taq DNA polymerase, nucleotides, and primers was inserted into a new set of microtubes. Next, 2 µL of each DNA sample was pipetted into their respective microtubes containing the master mix. The microtubes were then arranged in a thermal cycler to perform PCR reactions. PCR occurs in three stages: denaturing, annealing, and extending Figure 2. Denaturing is the process in which the double helix of DNA is separated into two strands. In the initial denaturation step, the temperature on the thermal cycler was raised to 94°C and held for 120 seconds. In all other denaturing instances, the temperature was held for only fifteen seconds. In the annealing steps, the temperature was lowered to 52°C and held for fifteen seconds to enable the primers to attach to respective DNA strands.
In the extending steps, the temperature was raised to 68°C to allow DNA polymerase to synthesize DNA strands complementary to the two template strands created in the denaturing step, thus replicating one segment of DNA into two identical segments The Polymerase Chain Reaction, 2024. Initially, a 400-base pair fragment of mitochondrial cytochrome b was copied in each sample. Later, an 800-base pair segment of mitochondrial cytochrome b was amplified in each sample using a different pair of primers. The DNA quality of each sample, including both the 400-base pair and 800-base pair versions, was visualized through gel electrophoresis (a technique used to separate DNA fragments according to their size) to determine which samples contained effective amplicons before delivery to Eurofins Genomics for sequencing.
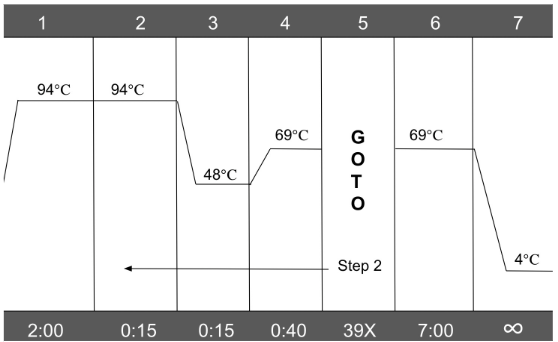
Figure 2:In order to amplify the 400 bp mitochondrial cytb sequence, PCR occurred in three stages: denaturing, annealing, and extending.
DNA Sequencing¶
Promising amplicons were sent for sequencing to Eurofins Genomics. Chromatogram reports of forward and reverse reads were received and aligned using Finch TV. Bases with quality scores below Q20 were not used in the process. Aligned sequences were blasted utilizing the Standard Nucleotide BLAST application on Genbank, and the closest matches were identified.
Results¶
In the first trial of the study, ten sequences precisely or closely aligned with the mitochondrial cytochrome b sequence of Sigmodon hispidus, more commonly known as the Hispid Cotton Rat. There were no other matches with the sequences in the database besides Homo sapiens and the only other result was unknown or unidentifiable, usually due to cross-contamination or lack of a strong DNA. Sigmodon hispidus is considered a species of least concern, according to The International Union for Conservation of Nature. They occupy habitats of heavily grassed areas such as fields and pastures, which are common in Morganton, North Carolina. They build nests under rocks or logs and create runways and tunnels both above and underground Copp, 2011. The basic local alignment search tool (BLAST) procedure for the first trial of the study resulted in the same species across all successfully sequenced samples.
Table 1:Ten samples matched closely with Hispid Cotton Rat
| Sample Number | Scientific Name | Percent Identity | Accession Number | Common Name |
|---|---|---|---|---|
| 1A | Sigmodon hispidus | 100.0% | AY263618.1 | Hispid Cotton Rat |
| 2A | Sigmodon hispidus | 99.38% | AF425205.1 | Hispid Cotton Rat |
| 4A | Sigmodon hispidus | 99.67% | AY263618.1 | Hispid Cotton Rat |
| 4B | Sigmodon hispidus | 100.0% | AY263618.1 | Hispid Cotton Rat |
| 4C | Sigmodon hispidus | 100.0% | AF425205.1 | Hispid Cotton Rat |
| 5A | Sigmodon hispidus | 100.0% | AF425205.1 | Hispid Cotton Rat |
| 6C | Sigmodon hispidus | 99.69% | AY263618.1 | Hispid Cotton Rat |
| 7D | Sigmodon hispidus | 100.0% | AY263618.1 | Hispid Cotton Rat |
| 7E | Sigmodon hispidus | 100.0% | AY263618.1 | Hispid Cotton Rat |
After running the study a second time, the identification of a second previously unidentified species as a food source for this particular owl occurred: Microtus pinetorum. The following images are photographs of the gels that were run. As shown below, the gels containing samples 4C and 4D resulted in one strong line and one light line, respectively. The third gel containing samples 2C and 2D heeded in two very light lines. The fourth gel had one strong line and three light lines, and gels one and five did not show up under the camera and light Figure 3. In the second trial of the study, only one out of the nine samples sequenced yielded sufficient base pairs to identify a match Table 2.
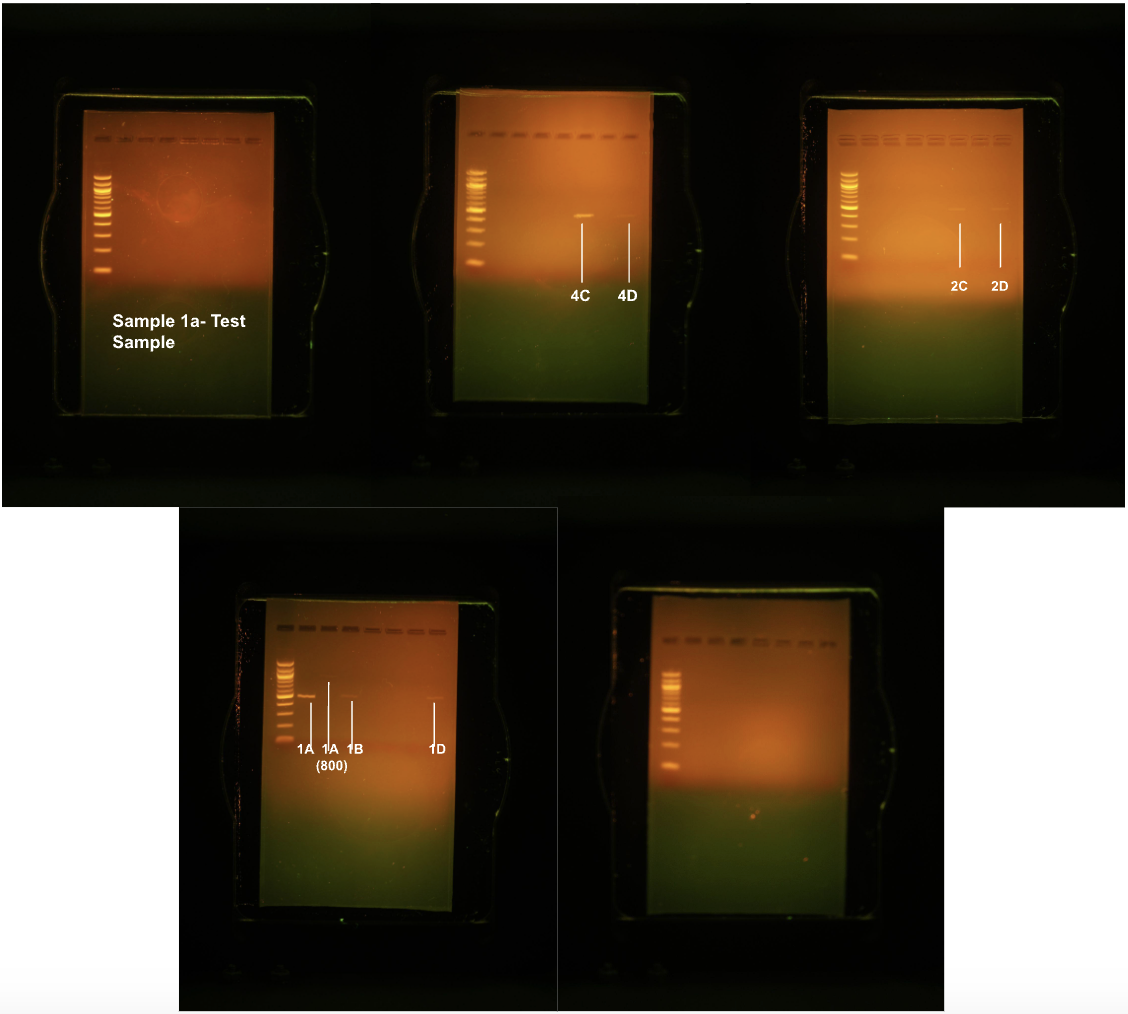
Figure 3:Gel electrophoresis during the second trial revealed that only eight DNA samples were of noticeable quality and worth sequencing.
Table 2:Sample 4C (containing 400 bp cytb) offered the greatest number of base pairs out of all sequenced samples to identify a match using Genbank’s database.
| Label | Trim | Q20 | Q20/Len | Q40 | Q40/Len |
|---|---|---|---|---|---|
| 1A_MVZ05 | 0 | 98 | 0.21 | 20 | 0.04 |
| 1A_MVZ04 | 0 | 1 | 0.01 | 0 | 0 |
| 1a_MVZ05 | 0 | 0 | 0 | 0 | 0 |
| 1a_MVZ16 | 0 | 2 | 0.02 | 0 | 0 |
| 1B_MVZ05 | 0 | 150 | 0.31 | 10 | 0.02 |
| 1B_MVZ04 | 0 | 170 | 0.52 | 19 | 0.06 |
| 1C_MVZ05 | 0 | 1 | 0.01 | 0 | 0 |
| 1C_MVZ04 | 5 | 0 | 0 | 0 | 0 |
| 1D_MVZ05 | 0 | 93 | 0.19 | 7 | 0.01 |
| 1D_MVZ04 | 0 | 23 | 0.05 | 0 | 0 |
| 2C_MVZ05 | 0 | 7 | 0.03 | 0 | 0 |
| 2C_MVZ04 | 0 | 68 | 0.24 | 0 | 0 |
| 2D_MVZ05 | 0 | 168 | 0.49 | 10 | 0.03 |
| 2D_MVZ04 | 112 | 235 | 0.51 | 43 | 0.09 |
| 4C_MVZ05 | 434 | 404 | 0.88 | 331 | 0.72 |
| 4C_MVZ04 | 375 | 405 | 0.87 | 370 | 0.8 |
| 4D_MVZ05 | 0 | 6 | 0.01 | 0 | 0 |
| 4D_MVZ04 | 0 | 109 | 0.11 | 0 | 0 |
DNA sequencing during the second trial of the study revealed a match with Microtus pinetorum, commonly known as the woodland vole, or the pine vole; it inhabits deciduous forests in eastern North America, living as a surface burrower. These voles are herbivorous, being a primary consumer in the food web of the Morganton ecosystem, and are an important source of food for many predators, including the owl that lived on the NCSSM Morganton campus, according to the DNA analysis results. Out of the sixteen samples used, only two came back with results that were possible to sequence. This may be due to the result of cross-contamination in the earlier parts of the DNA extraction process.
Discussion, Conclusions, Future Work¶
During the first round of the study, DNA sequencing results indicated that only one species, Sigmodon hispidus, was identified across the twenty-two samples of DNA found in the owl pellets. This finding disproved the initial claim that the samples would yield a diverse range of species. A potential error that could have led to this outcome might include cross-contamination among the samples, as indicated by the recurrence of the same two accession numbers (unique identifiers in sequence records) in the ten identified samples of Sigmodon hispidus. This presents a high possibility that contamination among the samples could have occurred at any point before the PCR process.
It is evident that DNA analysis led to the identification of owl prey at the species level, a level beyond the reach of morphological analysis, but it also introduced a risk of contamination that could potentially be mitigated through morphological analysis. The identification procedure yielded a success rate of approximately 45%, which is within the range of the success rates done in previous studies.
Sigmodon hispidus is known to be a carrier of respiratory diseases that can infect humans. Hantavirus Pulmonary Syndrome is a rare disease that is spread by Sigmodon hispidus and can be damaging to the human body. Specifically, the Black Creak hantavirus is spread by the cotton rat and has been identified in the Southeastern US in the warmer and more humid climate CDC, 2024. An airborne disease that is spread through feces, urine, and dander in areas of poor ventilation, such as small enclosed spaces with no ventilation. The disease can develop from a few days up to six weeks Hantavirus Infection, 2024. No treatment has been identified for this disease, making recovery longer and a higher death rate to high-risk patients.
Prevention is possible through disposing of rodents properly, such as identifying infestations, removing unwanted waste, and using proper equipment and solutions when cleaning, for instance gloves and 10% bleach solution to disinfect the area.
In the second trial of the study, sixteen samples were subjected to the analyzation process, and a second species, Microtus pinetorum, was identified among the DNA discovered in the selected owl pellets. This increases the initially known diversity of species from the samples by one hundred percent. The new species was identified from one specific sample and, upon sequencing, did not present any known contamination of human DNA, or cross-contamination with any other species, considering that the sequencing data did not show any significant outliers in the possible species. In addition, none of the remaining fifteen samples contained enough DNA to run a proper sequence and garner results that could indicate a certain species.
The rate of success in the second round of the study was significantly lower than that of the first analysis, possibly due to the idea that the pellets were approximately six months to a year older when analyzed than the initial twenty-two pellets were when they were analyzed. These pellets were collected at a later date, on top of the fact that it is unknown how long ago the pellets were formed, and the time between the formation of them and the collection of them. Over time, DNA begins to break down, and, if enough time has passed, can degrade to the point that running the PCR process on it is largely unsuccessful. The success rate of the second trial was approximately six percent, which is considerably lower than the range of success rates calculated in other studies. The explanation for the lower success rate than the previous trials is hypothesized to be due to the fact that the DNA from the more recently collected samples could have been more decayed than the initially collected samples, leading to a weaker sequencing process and therefore, fewer results.
Microtus pinetorum’s behavioral habit of surface burrowing directly affects the NCSSM campus, as parts of the campus are currently under construction. This burrowing can cause uneven or unstable ground, meaning that older buildings under construction may need their foundations strengthened or repaired to account for these changes. New buildings were constructed with these factors in mind, while the older buildings, constructed between the 1890s and early 1900s, may not have been. In addition, the woodland vole is also known to destroy crops and plants by stripping the bark from fruit trees, and feed on plant seeds or root systems Copp, 2011. This is economically harmful to the local farmers in the Morganton area who rely on their produce as a large source of income.
To enhance the diversity of species found in the pellet, the study could have included bird skulls identified in the owl pellet through a brief morphological analysis. This additional method would have provided a complementary perspective, offering a more comprehensive understanding of habitat life and the spread of disease in NCSSM Morganton. In the future, this method is intended to be used on other collected pellets, before the PCR process is performed on the samples. The process can also be used on samples dissected from the owl pellets that are not also being used for DNA analysis. In future trials, pellets will need to be collected from a different location and owl(s), due to the fact that the building the owl previously inhabited is now under construction, and the owl has relocated to an unknown location. This may aid in the discovery of more species, and increase the known biodiversity of species that the owls in Morganton, North Carolina consume.
We would like to thank Kovida Vupputuri for contributing to the experimental procedures during the first and second trials of the study, and for leading this study along with Sudiksha Battineni and Sophia Solomon.
Copyright © 2024 Battineni et al. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license, which enables reusers to distribute, remix, adapt, and build upon the material in any medium or format, so long as attribution is given to the creators.
- BLAST
- Basic Local Alignment Search Tool
- DNA
- Deoxyribonucleic acid
- NCSSM
- North Carolina School of Science and Mathematics
- PCR
- Polymerase Chain Reaction
- Wildlife Online. (2024). Owls | Wildlife Online. https://www.wildlifeonline.me.uk/animals/group/owls
- POULAKAKIS, N., LYMBERAKIS, P., PARAGAMIAN, K., & MYLONAS, M. (2005). Isolation and amplification of shrew DNA from barn owl pellets: BARN OWLS AND MICROMAMMALS. Biological Journal of the Linnean Society, 85(3), 331–340. 10.1111/j.1095-8312.2005.00500.x
- Rocha, R. G., Justino, J., Leite, Y. L. R., & Costa, L. P. (2015). DNA from owl pellet bones uncovers hidden biodiversity. Systematics and Biodiversity, 13(4), 403–412. 10.1080/14772000.2015.1044048
- Philips, J. R., & Dindal, D. L. (1979). Decomposition of raptor pellets. Journal of Raptor Research, 13(4), 2.
- Astral Scientific. (2024). 10 Questions you want to ask about Proteinase K. In Astral Scientific. https://astralscientific.com.au/blogs/astral-scientific/10-questions-you-want-to-ask-about-proteinase-k
- Afnan Uda, M. N., Parmin, N. A., Jambek, A. B., Hashim, U., Uda, M. N. A., & Shaharuddin, S. N. A. (2020). Evaluation and Optimization of Genomic DNA Extraction from Food Sample for Microfluidic Purpose. IOP Conference Series: Materials Science and Engineering, 743(1), 012031. 10.1088/1757-899x/743/1/012031
- The Polymerase Chain Reaction. (2024). [Training Material and Manuals]. https://www.nlm.nih.gov/ncbi/workshops/2022-10_Primer-BLAST/PCR.html
- Copp, D. (2011). Microtus pinetorum woodland vole. https://animaldiversity.org/accounts/Microtus_pinetorum/
- CDC. (2024). Preventing Hantavirus. https://www.cdc.gov/hantavirus/prevention/?CDC_AAref_Val=https://www.cdc.gov/hantavirus/hps/transmission.html
- Hantavirus Infection. (2024). https://www.nyc.gov/site/doh/health/health-topics/hantavirus-infection.page#:~:text=Peo
